Anonymous
7/17/2025, 9:07:12 AM No.17848943
https://m.economictimes.com/news/science/steppe-migration-to-india-was-between-3500-4000-years-ago-david-reich/amp_articleshow/71556277.cms
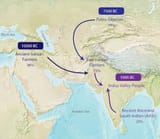
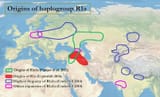
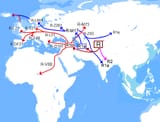
>Two recent papers- The Formation of Human Populations in South and Central Asia ( Vageesh Narasimhan et al) and An Ancient Harappan Genome lacks Ancestry from Steppe Pastoralists or Iranian Farmers (Vasant Shinde et al) - have sparked different interpretations on what they reveal about the genetics of ancient Indians. The papers were authored by a team of geneticists from Harvard Medical School working with Indian scientists. They studied ancient DNA from sites in Europe, Central Asia and South Asia, including a sample from the Indus Valley Civilisation site of Rakhigarhi, before drawing their conclusions. One of which was the contested claim that descendants of pastoralists from Eurasian steppes migrated to the Indian subcontinent in the first half of the second millenium BCE, "almost certainly" bringing Indo-European languages. Their studies also claim the Steppe migrants eventually contributed 0-30% of the genes of groups living in India today. In an email interview, Harvard geneticist Prof David Reich breaks down the findings. Excerpts
>1) What are the big takeaways from the 2 recent studies you co-authored - Vasant Shinde et al 2019, and Narasimhan et al 2019?
>1. At least some of the people of the ancient people of the Indus Valley Civilization were a mixture of south/southeast Asian-related hunter gatherers and Iranian-related hunter-gatherers. I say Iranian-related because their ancestors may actually have lived in South Asia rather than the Iranian plateau for many thousands of years before the time of the IVC. We just don’t know yet where they lived because of lack of ancient DNA from the relevant times and places
>Two recent papers- The Formation of Human Populations in South and Central Asia ( Vageesh Narasimhan et al) and An Ancient Harappan Genome lacks Ancestry from Steppe Pastoralists or Iranian Farmers (Vasant Shinde et al) - have sparked different interpretations on what they reveal about the genetics of ancient Indians. The papers were authored by a team of geneticists from Harvard Medical School working with Indian scientists. They studied ancient DNA from sites in Europe, Central Asia and South Asia, including a sample from the Indus Valley Civilisation site of Rakhigarhi, before drawing their conclusions. One of which was the contested claim that descendants of pastoralists from Eurasian steppes migrated to the Indian subcontinent in the first half of the second millenium BCE, "almost certainly" bringing Indo-European languages. Their studies also claim the Steppe migrants eventually contributed 0-30% of the genes of groups living in India today. In an email interview, Harvard geneticist Prof David Reich breaks down the findings. Excerpts
>1) What are the big takeaways from the 2 recent studies you co-authored - Vasant Shinde et al 2019, and Narasimhan et al 2019?
>1. At least some of the people of the ancient people of the Indus Valley Civilization were a mixture of south/southeast Asian-related hunter gatherers and Iranian-related hunter-gatherers. I say Iranian-related because their ancestors may actually have lived in South Asia rather than the Iranian plateau for many thousands of years before the time of the IVC. We just don’t know yet where they lived because of lack of ancient DNA from the relevant times and places
Replies: